News
CMC-GO-Term Activity: an R package for GO term activity transformation model
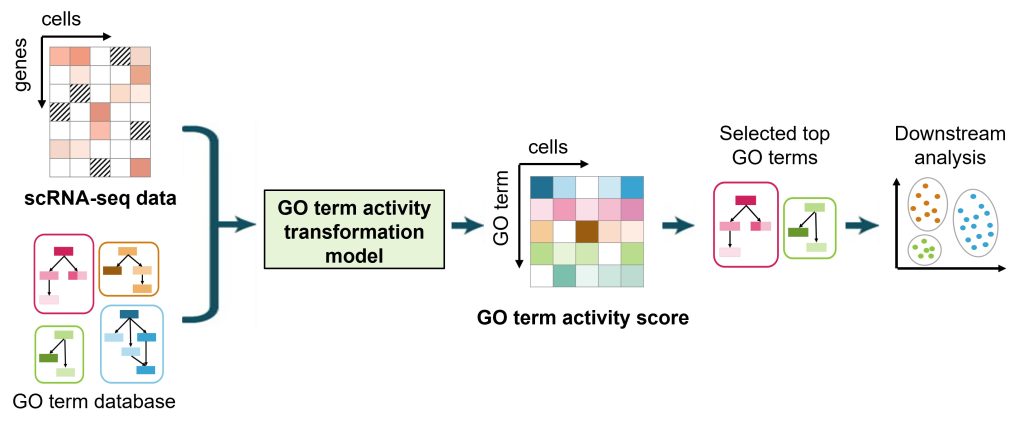
Single-cell RNA sequencing (scRNA-seq) technologies allow for the exploration of cellular and tissue heterogeneity. However, scRNA-seq data are highly noisy and suffer from dropout effects, which limit the power of these heterogeneity analyses. To overcome these challenges, Guoqiang Yu and Yue Wang’s groups developed a GO-term activity transformation model to transform scRNA-seq datasets into a GO-term activity score matrix. Since the GO-term activity score accounts for the expression of multiple functionally related genes, this score is more robust against noise and dropout effects.
Download and installation instructions: https://github.com/yu-lab-vt/CMC/tree/CMC-GOTermActivity
Find out more about CMC-GO-Term Activity: https://www.biorxiv.org/content/10.1101/2024.04.12.589306v1
CAM3.0: an R package for fully unsupervised deconvolution of complex tissues
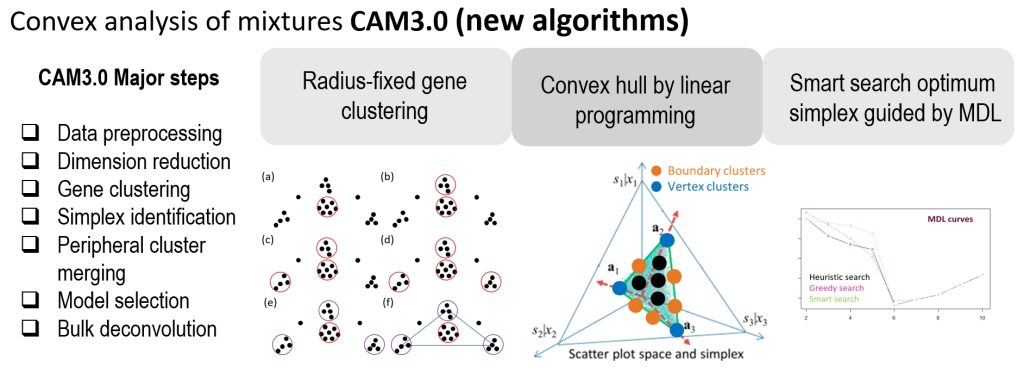
Complex tissues are dynamic ecosystems consisting of molecularly distinct yet interacting cell types. Computational deconvolution aims to dissect bulk tissue data into cell types and cell-specific information. However, most existing deconvolution tools exploit supervised approaches, requiring various types of references that may be unreliable or even unavailable for specific tissue microenvironments. CAM3.0 provides functions to perform fully unsupervised deconvolution on mixture expression profiles using Convex Analysis of Mixtures (CAM) and auxiliary tools to help interpret the cell type-specific results. Comparative experimental results obtained from both realistic simulations and case studies show that CAM3.0 can more accurately identify known or novel cell markers, determine cell proportions, and estimate cell-specific expressions from bulk tissue data.
Download and installation instructions: https://github.com/ChiungTingWu/CAM3?tab=readme-ov-file
Find out more about CAM3.0: https://pubmed.ncbi.nlm.nih.gov/38407991/
Jessica Moore was selected as a 2024 Schmidt Fellow
 Jessica Moore, a postdoctoral researcher in Cagla Eroglu’s laboratory at Duke University and a member of the A-Team group, has received a two-year fellowship from the prestigious Schmidt Science Fellows program. This initiative, established by Schmidt Sciences in 2018, aims to help researchers expand their work across various areas of study and build an interdisciplinary community of scientists dedicated to tackling bold challenges. As a 2024 Schmidt Science Fellow, Jessica will investigate the mechanisms governing the development of the gliovascular unit, a critical interface responsible for maintaining the integrity of the blood-brain barrier, particularly under stress conditions such as inflammation or neurodegenerative diseases. Specifically, she will study the impact of stressful stimuli on the brain-body axis, a critical first step toward developing interventions to reverse, halt, or mitigate the detrimental effects of stress on our body and brain.
Jessica Moore, a postdoctoral researcher in Cagla Eroglu’s laboratory at Duke University and a member of the A-Team group, has received a two-year fellowship from the prestigious Schmidt Science Fellows program. This initiative, established by Schmidt Sciences in 2018, aims to help researchers expand their work across various areas of study and build an interdisciplinary community of scientists dedicated to tackling bold challenges. As a 2024 Schmidt Science Fellow, Jessica will investigate the mechanisms governing the development of the gliovascular unit, a critical interface responsible for maintaining the integrity of the blood-brain barrier, particularly under stress conditions such as inflammation or neurodegenerative diseases. Specifically, she will study the impact of stressful stimuli on the brain-body axis, a critical first step toward developing interventions to reverse, halt, or mitigate the detrimental effects of stress on our body and brain.
The U19 A-Team extends its congratulations to Jessica!
Find out more here.
DDN3.0: An open-source software tool to determine significant rewiring of biological network structures
Complex diseases are often caused and characterized by the misregulation of multiple biological pathways. Differential network analysis enables joint inference of common and rewired biological network structures under different conditions. DDN3.0 provides basic functions to identify a network of significantly rewired molecular players potentially responsible for phenotypic transitions.
Find out more about DDN3.0: https://pubmed.ncbi.nlm.nih.gov/38902940/
Download and installation instructions: https://github.com/cbil-vt/DDN3
AQuA2 has been released! A fast, accurate, and versatile data analysis platform for the quantification of molecular spatiotemporal signals
 AQuA2 (Activity Quantification and Analysis 2) is a fast, accurate, and versatile data analysis platform built upon advanced machine learning techniques. AQuA2 allows for the quantification of spatiotemporal signals across biosensors, cell types, organs, animal models, and imaging modalities. Developed by Axel Nimmerjahn and Guoqiang Yu’s groups, AQuA2 is available for MATLAB and as a Fiji plugin.
AQuA2 (Activity Quantification and Analysis 2) is a fast, accurate, and versatile data analysis platform built upon advanced machine learning techniques. AQuA2 allows for the quantification of spatiotemporal signals across biosensors, cell types, organs, animal models, and imaging modalities. Developed by Axel Nimmerjahn and Guoqiang Yu’s groups, AQuA2 is available for MATLAB and as a Fiji plugin.
Find out more about AQuA2: https://www.biorxiv.org/content/10.1101/2024.05.02.592259v1
Download and installation instructions: https://github.com/yu-lab-vt/AQuA2?tab=readme-ov-file
The Salk Institute promotes neuroscientist Axel Nimmerjahn
 In recognition of his notable and innovative contributions to science, the Salk Institute’s Axel Nimmerjahn, PhD, has been promoted from associate professor to full professor. Axel Nimmerjahn leads the U19 A-Team Program and is the director of Salk’s Waitt Advanced Biophotonics Center. With his lab, he has developed and applied new tools to visualize cells’ structural and functional dynamics in the brain and spinal cord under healthy and diseased conditions, including tiny wearable microscopes allowing scientists to observe cellular activities and molecular signaling in real-time in behaving animals. The Nimmerjahn lab’s research focuses on brain and spinal cord function, neuro-immune interactions, and diseases such as Alzheimer’s disease, neuroinflammation, spinal cord injury, brain cancer, stroke, and pain.
In recognition of his notable and innovative contributions to science, the Salk Institute’s Axel Nimmerjahn, PhD, has been promoted from associate professor to full professor. Axel Nimmerjahn leads the U19 A-Team Program and is the director of Salk’s Waitt Advanced Biophotonics Center. With his lab, he has developed and applied new tools to visualize cells’ structural and functional dynamics in the brain and spinal cord under healthy and diseased conditions, including tiny wearable microscopes allowing scientists to observe cellular activities and molecular signaling in real-time in behaving animals. The Nimmerjahn lab’s research focuses on brain and spinal cord function, neuro-immune interactions, and diseases such as Alzheimer’s disease, neuroinflammation, spinal cord injury, brain cancer, stroke, and pain.
Read the full story: https://www.salk.edu/news-release/the-salk-institute-promotes-neuroscientist-axel-nimmerjahn/.
Cagla Eroglu elected to American Academy of Arts & Science
 Cagla Eroglu, PhD, who is leading the U19 A-Team Program’s effort to uncover the role of astrocytes in goal-oriented behaviors, is one of the investigators elected to the American Academy of Arts & Sciences for 2024. The academy’s current members represent innovative thinkers in every field and profession. The 250 members elected in 2024 are being recognized for their excellence and invited to uphold the Academy’s mission of engaging across disciplines and divides.
Cagla Eroglu, PhD, who is leading the U19 A-Team Program’s effort to uncover the role of astrocytes in goal-oriented behaviors, is one of the investigators elected to the American Academy of Arts & Sciences for 2024. The academy’s current members represent innovative thinkers in every field and profession. The 250 members elected in 2024 are being recognized for their excellence and invited to uphold the Academy’s mission of engaging across disciplines and divides.
Read the full story: https://medschool.duke.edu/news/eroglu-elected-american-academy-arts-sciences.
Daniel Quintero awarded with prestigious NSF Fellowship
 Daniel Quintero, a graduate student in Cagla Eroglu’s laboratory at Duke University and a member of the A-Team group, has received a five-year fellowship from the NSF Graduate Research Fellowship Program (GRFP). Daniel is actively investigating the cellular and molecular mechanisms by which astrocytes, the star-shaped glial cells of the brain, influence sleep. The U19 A-Team Leadership extends its congratulations to Daniel!
Daniel Quintero, a graduate student in Cagla Eroglu’s laboratory at Duke University and a member of the A-Team group, has received a five-year fellowship from the NSF Graduate Research Fellowship Program (GRFP). Daniel is actively investigating the cellular and molecular mechanisms by which astrocytes, the star-shaped glial cells of the brain, influence sleep. The U19 A-Team Leadership extends its congratulations to Daniel!
More information: https://medschool.duke.edu/news/duke-graduate-students-awarded-prestigious-nsf-fellowships.
BILCO: An Efficient Algorithm for Joint Alignment of Time Series
 BILCO (BIdirectional pushing with Linear Component Operations) is an efficient algorithm, developed by Guoqiang Yu’s group at Virginia Tech, for solving joint alignment problems of time series and min-cut for GTW (graphical time warping) graphs. BILCO has the same theoretical time complexity as the best popular methods, such as HIPR (highest-label push-relabel), but it provides a significant empirical efficiency boost without sacrificing the accuracy of joint alignment. On thousands of datasets under various simulated scenarios and real application cases (calculate signal propagation calculation, exact depth information extraction, and signature identification), BILCO shows around 10 to 50 times speed improvement and only cost 1/10 memory compared with the best peer methods IBFS (incremental breadth-first search), HPF (Hochbaum’s pseudo flow), BK (Boykov-kolmogorov), and HIPR.
BILCO (BIdirectional pushing with Linear Component Operations) is an efficient algorithm, developed by Guoqiang Yu’s group at Virginia Tech, for solving joint alignment problems of time series and min-cut for GTW (graphical time warping) graphs. BILCO has the same theoretical time complexity as the best popular methods, such as HIPR (highest-label push-relabel), but it provides a significant empirical efficiency boost without sacrificing the accuracy of joint alignment. On thousands of datasets under various simulated scenarios and real application cases (calculate signal propagation calculation, exact depth information extraction, and signature identification), BILCO shows around 10 to 50 times speed improvement and only cost 1/10 memory compared with the best peer methods IBFS (incremental breadth-first search), HPF (Hochbaum’s pseudo flow), BK (Boykov-kolmogorov), and HIPR.
More information about BLICO: https://github.com/yu-lab-vt/BILCO.
Congrats to the U19 Astrocyte-Team!
 The U19 Astrocyte-Team (A-Team) members have made significant advancements in their field, resulting in a total of 14 awards and fellowships in the first two years of the U19 A-Team Program. The U19 A-Team Leadership extends their congratulations for their remarkable accomplishments. They also acknowledge and appreciate the invaluable contribution of the A-Team Scholars in expanding our understanding of the crucial roles played by astrocytes in neural circuit operation, complex behaviors, and brain computation.
The U19 Astrocyte-Team (A-Team) members have made significant advancements in their field, resulting in a total of 14 awards and fellowships in the first two years of the U19 A-Team Program. The U19 A-Team Leadership extends their congratulations for their remarkable accomplishments. They also acknowledge and appreciate the invaluable contribution of the A-Team Scholars in expanding our understanding of the crucial roles played by astrocytes in neural circuit operation, complex behaviors, and brain computation.
Recipients from the four institutions part of the U-19 A-Team are listed here.
Synbot: An open-source image analysis software for automated quantification of synapses
 The precise formation of neuronal connections, known as synapses, is essential for proper brain function. As a result, understanding the mechanisms of synaptogenesis has been a major focus in cellular and molecular neuroscience. Immunohistochemistry, coupled with light microscopy, is commonly used to label and visualize synapses. However, quantifying the number of synaptic contacts from light microscopy images has traditionally been a challenging and time-consuming task, with variable results between experimenters.
The precise formation of neuronal connections, known as synapses, is essential for proper brain function. As a result, understanding the mechanisms of synaptogenesis has been a major focus in cellular and molecular neuroscience. Immunohistochemistry, coupled with light microscopy, is commonly used to label and visualize synapses. However, quantifying the number of synaptic contacts from light microscopy images has traditionally been a challenging and time-consuming task, with variable results between experimenters.
To overcome these limitations, Cagla Eroglu’s group at Duke University has developed SynBot, a new open- source Image-based software. SynBot addresses the technical bottlenecks of traditional synapse quantification analysis by automating several stages of the process and incorporates the machine learning algorithm ilastik, which enables accurate thresholding for synaptic puncta identification. Additionally, the software’s code is easily modifiable, allowing users to adapt it to their specific needs.
Find out more about Synbot: https://pubmed.ncbi.nlm.nih.gov/37425715/
Download and installation instructions: https://github.com/Eroglu-Lab/Syn_Bot
Check out AQuA (Astrocyte Quantification and Analysis), a tool to detect signaling events
AQuA (Astrocyte Quantification and Analysis) is an effective tool to detect signaling events from microscopic time-lapse imaging data of astrocytes or other cell types. The algorithm, developed by Guoqiang Yu’s group at Virginia Tech, is data-driven and based on machine learning principles and can be applied across model organisms, fluorescent indicators, experimental modalities, cell types, and imaging parameters.
More information about AQuA: https://github.com/yu-lab-vt/AQuA
A prototype for a web-based service version of AQuA is also under development. Please stay tuned!